Recombinant PNGase F
LudgerZyme recombinant PNGase F kit (New England BioLabs).
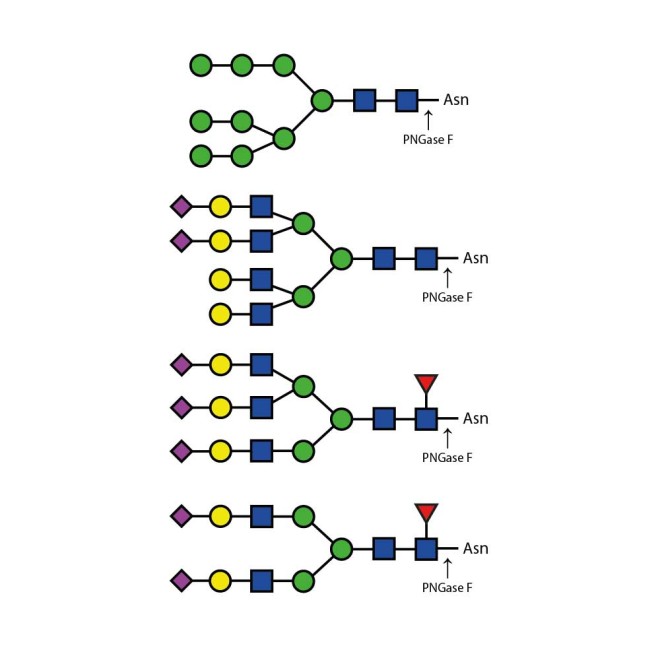
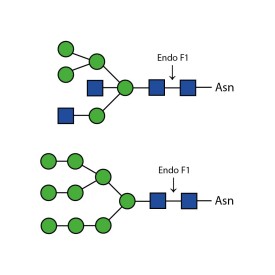
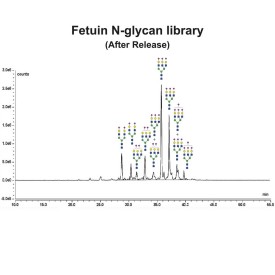
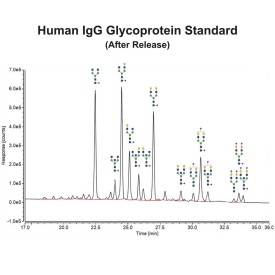
PNGase F is suitable for the release of all types (high-mannose, hybrid and complex) N-linked glycans from glycoproteins and glycopeptides. PNGase F will not remove oligosaccharides containing α(1-3) linked core fucose commonly found on plant glycoproteins.
LudgerZyme PNGase F is a recombinant glycosidase cloned from Elizabethkingia miricola and expressed in E. coli. The enzyme is supplied glycerol-free (for optimal performance in HPLC-intensive methods) along with Reaction Buffer, Denaturation Solution and NP-40 Solution for efficient de-glycosylation.
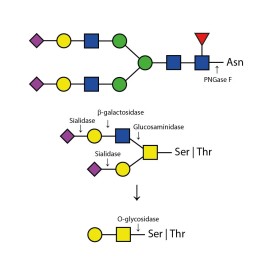
LudgerZyme Peptide N-glycosidase F (PNGase F) is suitable for the release of N-linked glycans in solution and from immobilized samples. The enzyme cleaves between the innermost GlcNAc of the oligosaccharide moiety at its attachment point to the asparagine residue on the protein and subsequently converts the asparagine into aspartic acid. Released glycans with free reducing terminus can be labelled using LudgerTag labelling technology for fluorescence and high MS sensitivity detection.Kit includes enzyme plus reaction buffers. Sufficient for up to 150 reactions.
Note: The LZ-rPNGaseF-30 enzyme kit part of the LT-KAB-VP30-MOD contains enzyme plus reaction buffers that is sufficient for 30 reactions.
Product specification:
LudgerZyme PNGase F is a recombinant glycosidase cloned from Elizabethkingia miricola and expressed in E. coli. The enzyme is supplied glycerol free (for optimal performance in HPLC intensive methods) along with Reaction Buffer, Denaturation Solution and NP-40 Solution for efficient de-glycosylation.
Application:
LudgerZyme Peptide N-glycosidase F (PNGase F) is suitable for release of N-linked glycans in solution, and from immobilized samples. The enzyme cleaves between the innermost GlcNAc of the oligosaccharide moiety at its attachment point to the asparagine residue on the protein and subsequently converts the asparagine into aspartic acid. Released glycans with free reducing terminus can be labelled using LudgerTag labelling technology for fluorescence and high MS sensitivity detection.
Source: Elizabethkingia miricola in E. coli
EC: 3.5.1.52
Contents:
PNGase F (Elizabethkingia miricola) suuplied in 50 mM NaCl 5 mM EDTA 20 mM Tris-HCl pH 7.5 - 1 vial of 0.15mL
10X Reaction Buffer 500 mM sodium phosphate (pH 7.5 at 1X dilution) - 1 vial of 1.0 mL
10X Denaturation Solution 5% SDS 400 mM DTT - 1 vial of 1.0 mL
NP-40 10% solution - 1 vial of 1.0 mL
Suggested Usage:
Denaturing reaction conditions:
1. Make up sample volume to 9 µL with ultrapure water.
2. Add 1 µL of 10X Denaturation Solution to each glycoprotein sample. Close the reaction vials, vortex thoroughly and briefly centrifuge to ensure the samples are completely dissolved.
3. Incubate the samples at 100°C for 10 minutes.
4. Add 2 µL of 10X Reaction Buffer to each glycoprotein sample.
5. Add 2 µL of 10% NP-40 solution.
6. Adjust the reaction volume to 20 µL by adding 6 µL of water.
7. Add 1 µL of PNGase F. Close the reaction vials, mix gently and briefly centrifuge.
8. Incubate the samples at 37°C for 1h.
Specificity:
PNGase F is suitable for release of all types (high-mannose, hybrid and complex) N-glycans from glycoproteins and glycopeptides. Xaa-Asn-Xaa sequence is the minimal peptide substrate for this enzyme. Note that some non-mammalian glycans from sources such as plants, insects and parasites carrying α1-3 linked core fucose will not be cleaved with PNGase F. For these samples PNGase A can be used.
Number of Samples: Kit contains 75,000 units of PNGase F at concentration of 500,000 units/ml. Sufficient for approximately 150 samples
Amount of Samples: As a guideline up to 100 µg of glycoprotein per sample
Suitable Samples: Glycoproteins and glycopeptides containing N-linked glycans
Storage:
Store at 4°C. Protect from sources of heat and light
Heat Inactivation:
PNGase F is inactivated after 10 minutes at 75°C.