Enzymatic DeGlycoMx Kit
References:
1. Kobata, A. Use of endo- and exoglycosidases for structural studies of glycoconjugates. Anal Biochem 100:1- 14 (1979).
2. Kim MS, Leahy D. Enzymatic deglycosylation of glycoproteins. Methods Enzymol. 533:259-63 (2013).
3. Magnelli PE, Bielik AM, Guthrie EP. Identification and characterization of protein glycosylation using specific endo- and exoglycosidases. J Vis Exp. Dec 26;(58):e3749 (2011).
4. Segu ZM, Hussein A, Novotny MV, Mechref Y. Assigning N-glycosylation sites of glycoproteins using LC/MSMS in conjunction with endo-M/exoglycosidase mixture. J Proteome Res. Jul 2;9(7):3598-607 (2010).
5. Sojar, H. T. and O.P. Bahl. A chemical method for the deglycosylation of proteins. Arch Biochem Biophys 259:52-57 (1987).
6. Tarentino A.L. and T.H. Plummer. Enzymatic deglycosylation of asparagine- linked glycans: purification, properties, and specificitly of oligosaccharide- cleaving enzymes from Flavobacterium meningosepticum. Methods in Enzymol 230: 44-57 (1994).
7. Uchida, Y., Y. Tsukada and T. Sugimori. Enzymatic properties of neuraminidases from Arthrobacter ureafaciens. J Biochem (Tokyo) 86:573-585 (1979).
8. Zhang W, Wang H, Zhang L, Yao J, Yang P. Large-scale assignment of N-glycosylation sites using complementary enzymatic deglycosylation. Talanta. Jul 15;85(1):499-505 (2011).
This kit developed for protein deglycosylation includes our DeGlycoMx, a premixed cocktail of the enzymes required to remove all N-linked oligosaccharides and most O-linked sugars from 0.5 mg of glycoprotein, via 10 reactions of up to 50 micrograms of protein per reaction.
The kit is easy-to-use and effective: add 2 µL of the DeGlycoMx enzyme to your denatured sample and the included buffer and incubate for 3 hours at 37°C.
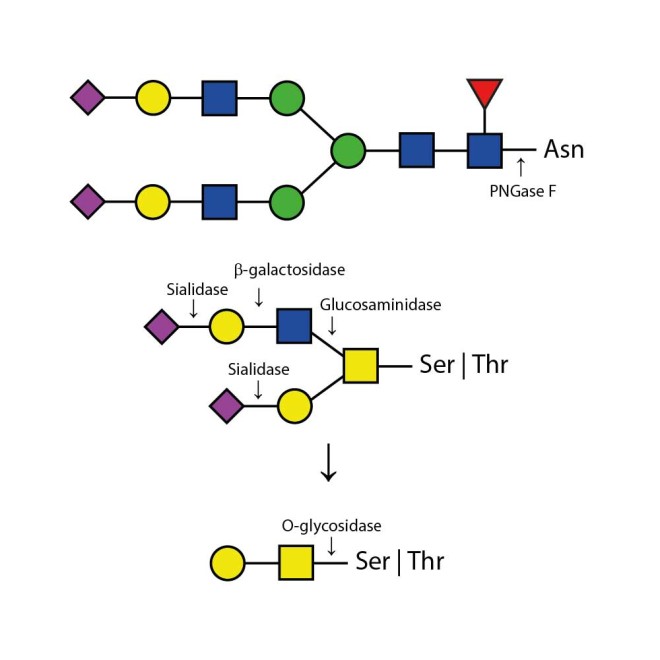
The enzymes are provided as one 20 µL premixed cocktail including PNGase F (Elizabethkingia meningosepticum), O-Glycosidase (Streptococcus pneumoniae), Sialidase (Arthrobacter ureafaciens), β-Galactosidase (Streptococcus pneumoniae), Glucosaminidase (Streptococcus pneumoniae).
Kit includes enzyme plus reaction buffers. Sufficient for up to 20 reactions.
Product specification:
The enzymes are premixed in either one 20 or 100 microliter vial. This allows researchers to quickly remove most all glycans (with the exception of O-Linked mucins) from their glycoprotein in both denatured and native conditions. By combining the enzymes into one easy-to-use premixed solution, researchers save time and money in the pursuit of protein deglycosylation. The enzymes can also be supplied individually in 20 microliter vials of each enzyme (KE-DG01) allowing the researcher to the flexibility to characterize the glycans attached to their glycoprotein more fully than with our mixture of enzymes.
Contents:
The enzymes are provided as one 20 µL premixed cocktail including: PNGase F (Elizabethkingia meningosepticum), O-Glycosidase (Streptococcus pneumoniae), Sialidase (Arthrobacter ureafaciens), β-Galactosidase (Streptococcus pneumoniae), Glucosaminidase (Streptococcus pneumoniae).
Also includes:
5x Reaction buffer - 200 µL
Denaturation Solution - 100 µL
Triton X - 100 µL
Specificity:
The Enzymatic DeGlycoMx Kit will remove all N-linked oligosaccharides and many O-linked oligosaccharides from glycoproteins. Protein deglycosylation for N-linked glycans (Asparganine-linked) is performed using the enzyme PNGase F. In addition, all Serine or Threonine linked (O-linked) Gal-(β1-3)-GalNAc-(α1) and all sialic acid substituted Gal-(β1-3)-GalNAc-(α1) will be removed using the combination of Sialidase and O-Glycosidase. The addition of β-Galactosidase and Glucosaminidase will assist in the deglycosylation of larger O-link structures.
Directions:
1. Mix 10 µL of reaction buffer with up to 50 µg of glycoprotein in 33 µL distilled water in a 1.5 µL tube.
2. Add 2.5 µL denaturation solution. Mix gently and place in boiling water bath for 5 minutes. Chill on ice.
3. Add 2.5 µL of Triton-X.
4. Add 2 µLs of the DeGlycoMx enzyme cocktail. Incubate for 3 hours at 37°C.
Note: Denaturation increases the rate of enzyme digestion up to 10 fold. If denaturation is not desired omit step 2-3, add with 5 µL of distilled water and increase incubation time up to 24 hours.