β-N-acetylglucosaminidase
References:
1. Clarke, V. A., N. Platt and T.D. Betters. Cloning and expression of the beta-N-acetylglucosaminidase gene from Steptococcus pneumoniae. Generation of truncated enzymes with modified aglyconn specificity. J Biol Chem 270:8805-8814 (1995).
2. Dwek, R. A., C. J. Edge, D. J. Harvey, M. R. Wormald and R. B. Parekh. Analysis of glycoprotein-associated oligosaccharides. Ann Rev Biochem 62: 65-100 (1993).
3. Glasgow, L.R., J.C. Paulson and R.L. Hill. Systematic purification of five glycosidases from Streptococcus pneumoniae J Biol Chem 252:8615-8623(1977).
4. Kobata, A. Use of endo- and exoglycosidases for structural studies of glycoconjugates. Anal Biochem 100: 1-14 (1979).
5. Prime, S., J. Dearnley , A. M. Venton, R. B. Parekh and C. J. Edge. Oligosaccharide sequencing based on exo- and endoglycosidase digestion and liquid chromatographic analysis of the products. J Chromatogr A 720: 263-274 (1996).
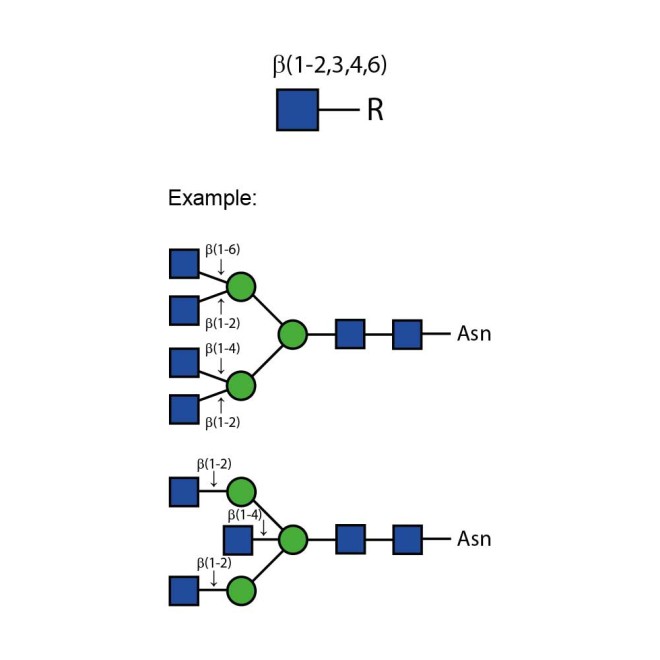
N-acetylglucosaminidase cleaves all non-reducing terminal β-linked N-acetylglucosamine residues from complex carbohydrates and glycoproteins.
The cleavage rates of different linkages of GlcNAc on bi-, tri- and tetraantennary oligosaccharides is greatly dependent on the steric hindrance by neighbouring residues. The β(1-2)GlcNAc residue linked to the β(1-3)-linked mannose is cleaved at the highest rate and the β(1-2) GlcNAc residue linked to the β(1-6)-linked mannose at the lowest rate for all three oligosaccharides. The β(1-6) GlcNAc residue, when present, is removed at the second highest rate and the β(1-4) GlcNAc, third. On a triantennary structure, this residue is removed at the second highest rate. A bisecting β(1-4) GlcNAc linked to the β-linked mannose severely hinders cleavage of other GlcNAc residues–high concentrations of enzymes and prolonged incubation times are required for cleavage.
Product specification
Source: Recombinant from Streptococcus pneumoniae in E. Coli.
EC: 3.2.1.52
Alternate Names: β-N-acetylglucosaminidase, N-acetyl-β-d-glycosaminide N-acetylglucosaminohydrolase, glucosaminidase, hexosaminidase
Contents:
-n-acetylglucosaminidase-kit.jpg)
N-acetylglucosaminidase in 20 mM Tris-HCl, 25 mM NaCl, pH 7.5
5x Reaction Buffer 250 mM sodium phosphate, pH 5.0
Specific Activity: >80 U/mg
Activity: >50 U/mL
Molecular weight: ~140,000 daltons
pH range: 5-7, optimum 5.0
Suggested usage:
1. Add up to 100 µg of glycoprotein or 1 nmole of oligosaccharide to a tube.
2. Adjust to 14 µL final volume with de-ionized water.
3. Add 4 µL 5x reaction buffer (pH 5.0).
4. Add 2 µL of N-acetylglucosaminidase.
5. Incubate 3 hours at 37°C.
NOTE: If bisecting GlcNAc or β(1-2)GlcNAc-α(1-6)Man are present increase incubation nation time to 18 hours.
Specificity: Cleaves all non-reducing terminal β-linked N-acetylglucosamine. Bisecting GlcNAc slows the reaction.
Specific Activity Assay: Defined as the amount of enzyme required to produce 1 µmole of p-nitrophenol (pNP) in 1 minute at 37°C, pH 5.0 from p-nitrophenyl-β-D-N-acetyl-glucosaminide
Storage: Store enzyme at 4°C.
Purity: Each lot of N-acetylglucosaminidase is tested for contaminating protease as follows; 10 µg of denatured BSA is incubated for 24 hours with 2 µL of enzyme. SDS-PAGE analysis of the treated BSA shows no evidence of degradation.
The production host strain has been extensively tested and does not produce any detectable glycosidases.
Stability: Stable at least 12 months when stored properly. Several days exposure to ambient temperatures will not reduce activity.















-275x275.jpg)
-275x275.jpg)
-275x275.jpg)